Projects
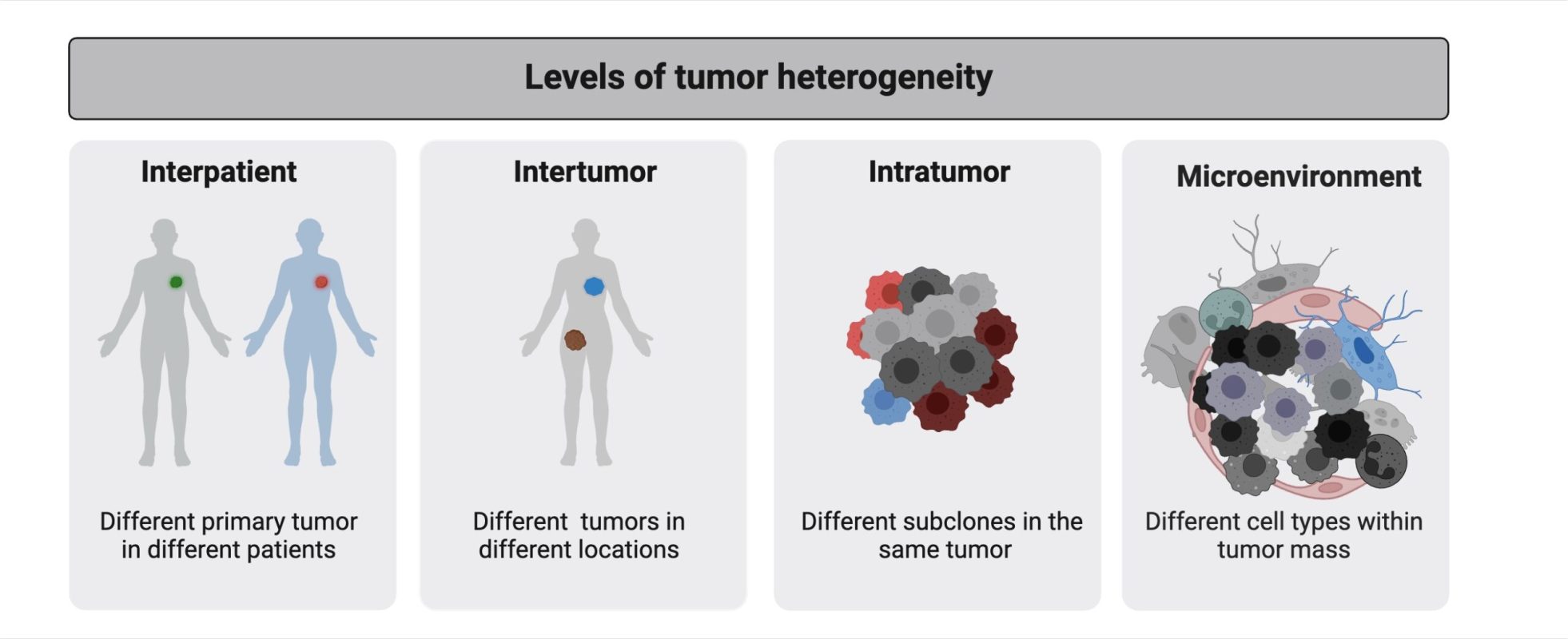
1. Tumor heterogeneity is present at all levels of cancer progression.
Cancer is inherently heterogeneous, evolving through genetic and cellular diversity that fuels therapy resistance and disease progression. This variability exists at multiple levels – between patients, within tumors, and even among individual cancer and stromal cells. As a result, no single treatment is universally effective, and tumors often relapse despite initial therapy response. Understanding this complexity underscores the need for personalized treatment strategies to improve patient outcomes.
2. Cancer evolution in colorectal cancer
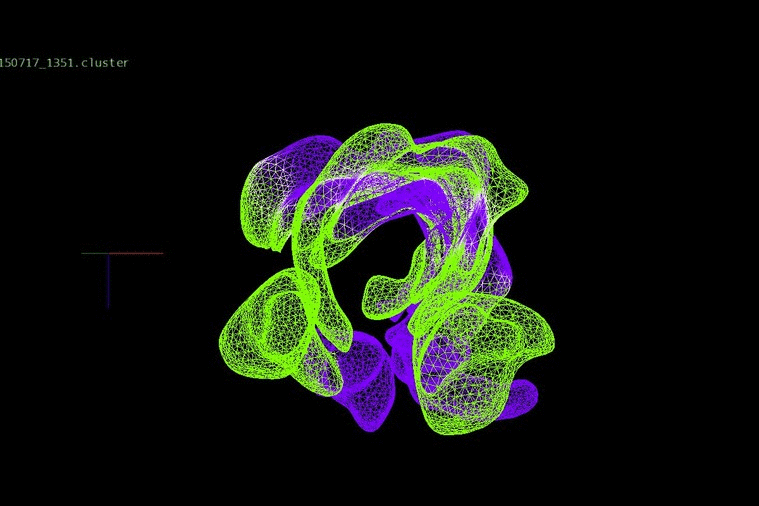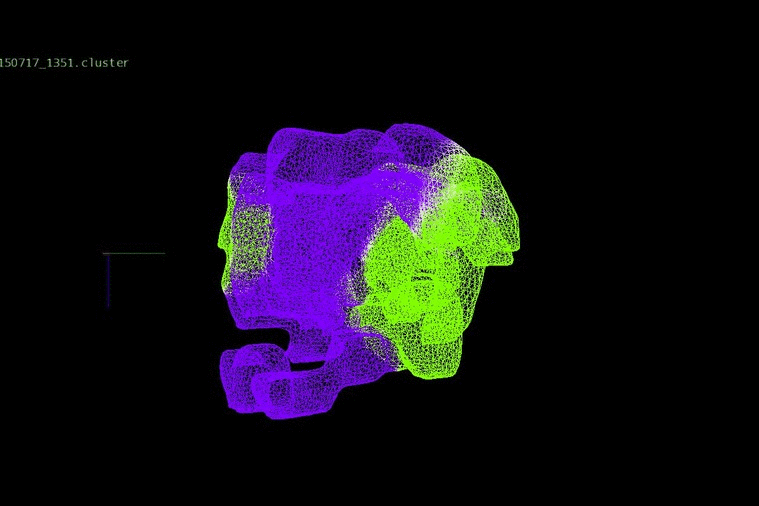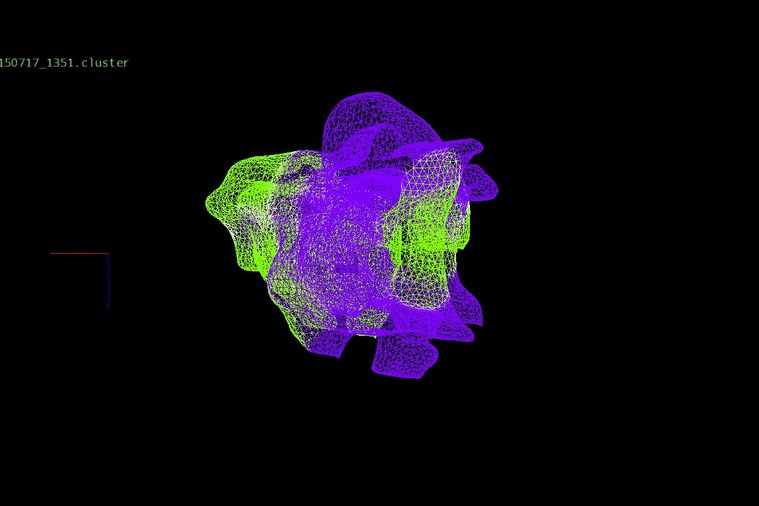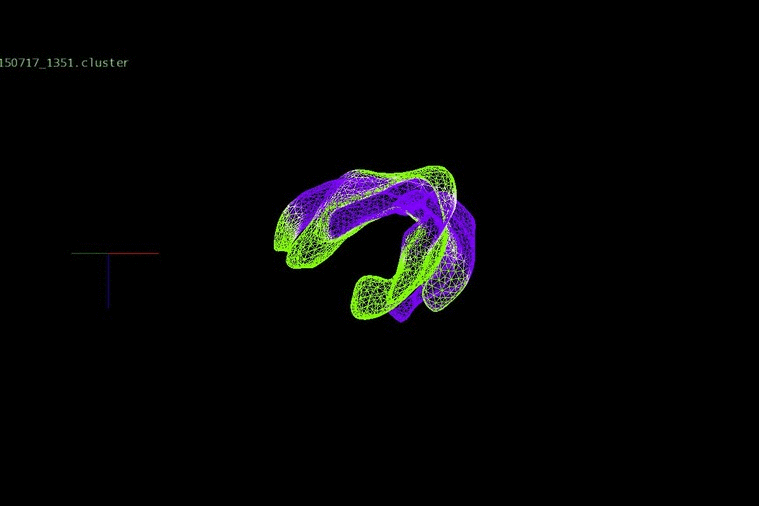
DNA heterogeneity can identify changes in adult cancers, which, when placed together in an evolutionary concept, expose relationships between clones within a patient’s disease. We have been investigating evolution and progression in colorectal cancer from the early stages of adenomas to late stages of liver metastases and relapse.
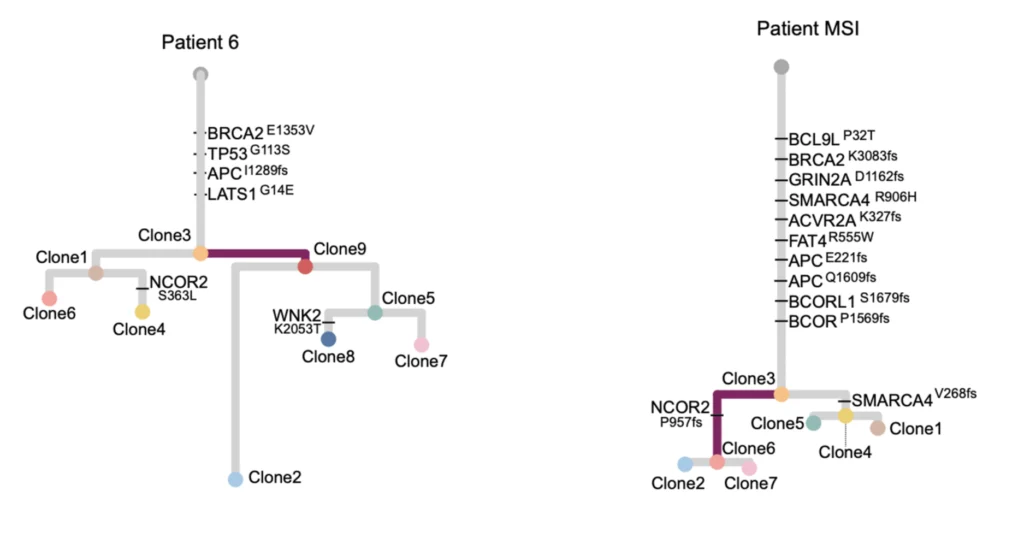
We identified DNA copy number heterogeneity in one untreated colorectal cancer, which was divided into 68 different pieces and reconstructed into a 3D representation of its original histology (above). A phylogenetic representation of two colorectal cancer patients with liver metastases and their relapse (purple) in a study with Dr. Laura Tomás from D. Posada Lab (below).
3. Cancer evolution in neuroblastoma

High-risk, treatment-refractory and relapsed neuroblastomas are among the most lethal childhood cancers. Outcomes are difficult to predict and relapses are frequent and despite aggressive multimodal treatment more than half of patients with high-risk disease relapse, often within two years of diagnosis. This therapy failure is largely driven by the tumor’s ability to adapt. Using patient-derived tumor models, we are looking at subpopulations that evolve spatially and temporally under therapeutic pressure.
4. Building patient-derived tumoroid platforms: mimicking, monitoring, and steering cancer evolution
Patient-derived tumoroids are 3D ex vivo models of a patient’s tumor.
Our team explores the evolving subclonality of sensitive and resistant cells, assesses their fitness costs and examines various selective treatment responses.
We have established a patient-derived tumoroid platform which enables genetic and fluorescent tracing of resistant and sensitive populations in experiments exploring tumor evolution and its molecular mechanisms The platform creates, combines and tracks resistant and sensitive cell populations to investigate evolutionary concepts that can inform future treatment strategies using evolutionary dynamics.
5. Plasticity in treatment and relapse
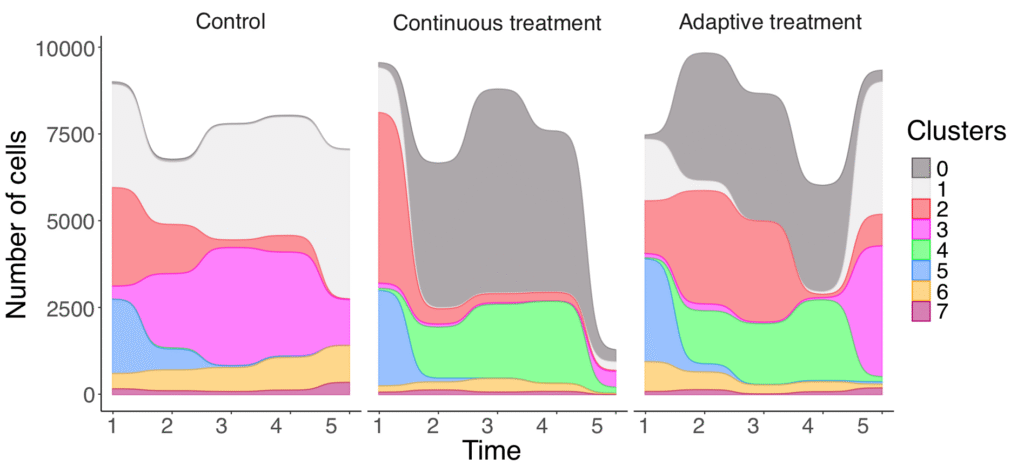
Cancer plasticity is an important mechanism of treatment resistance and relapse. We use single-cell RNA data from tumor models to investigate how this plasticity is shaped over time. In the graphical visualization, we follow cluster movements from patient-derived organoids under different treatment schemes.